Unveiling How Coronavirus Hijacks Our Cells to Help Rush New Drugs to Patients
UCSF-led Rapid-Response Research Team Takes an Unconventional Approach to Fighting Viruses
UC San Francisco’s Nevan Krogan, PhD, hasn’t slept much since January, when cases of COVID-19 caused by a novel coronavirus began to spike in China, stoking fears of the ensuing global pandemic that has since affected more than 100 countries.
During those fateful weeks, Krogan, director of UCSF’s Quantitative Biosciences Institute (QBI) and professor of Cellular and Molecular Pharmacology, has been single-mindedly focused on one goal: understanding in depth how the SARS-CoV-2 virus that causes COVID-19 hijacks the machinery of human cells to perpetuate itself. That knowledge could lead the way to stopping the disease in its tracks.
Though many hopes are hanging on the development of a vaccine or drug that targets the virus directly, Krogan takes an unconventional approach: target the host – in other words, you.

Nevan Krogan (center), PhD, and his collaboration of scientists around the world have fast-tracked efforts to find the proteins in cells that get highjacked by COVID-19. Photo by Susan Merrell
Considering exactly how a virus infects its host reveals why Krogan, also a senior investigator at the UCSF-affiliated Gladstone Institutes, takes aim from this unusual direction. Viruses have great power to cause illness, but they also have a critical weakness: they’re unable to reproduce on their own and depend on the protein-making machinery of their hosts to propagate. It’s that weakness that Krogan sees as an efficient means to put the brakes on the runaway train of COVID-19, and maybe other diseases as well.
To infect us, the SARS-CoV-2 virus gets its genetic material into our cells and then co-opts our own proteins, reassigning them to the task of making millions of copies of itself. This tsunami of copied viruses ultimately kills cells, releasing virus particles that travel through the body to infect more cells or be spread to new human hosts, perpetuating the outbreak.
Hunting for the Protein Interactions
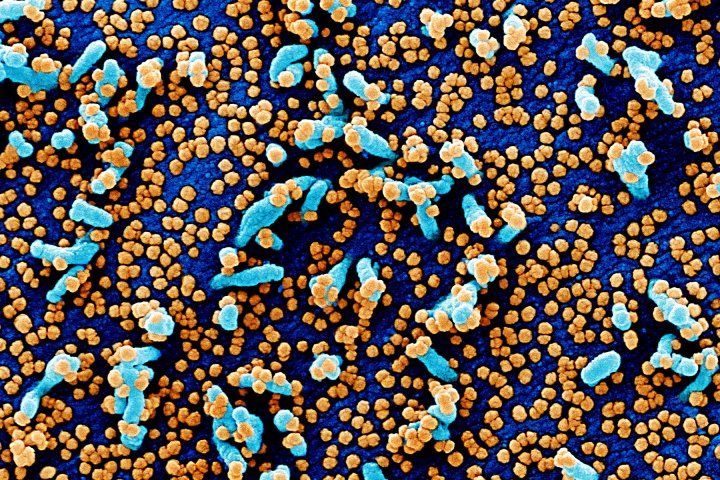
A colorized scanning electron micrograph shows cultured cells (blue) heavily infected with SARS-COV-2 virus particles (orange). Image by NIH
After a round-the-clock, two-month process, Krogan identified more than 300 human proteins that interact with SARS-CoV-2 during infection. To do this, members of his lab looked at all 29 genes in the SARS-CoV-2 genome and sorted out the protein products of 26 of them. Then they cloned these proteins, added tiny chemical tags to them, and put them in human cells. The human proteins that interacted with viral ones could be identified by the chemical tags. The team compiled this information into a library of human proteins that enable the virus to replicate in our bodies.
Based on this information, Krogan and colleagues made a map of this “interactome,” laying out a network diagram showing how our proteins cooperate with viral ones. Blocking these interactions may prevent the virus from making copies of itself, Krogan reasons, thereby slowing down both infection and transmission.
Searching for Drugs
To identify the drugs that might accomplish this end, in a matter of days Krogan assembled a scientific coalition, dubbed the QBI Coronavirus Research Group, or QCRG, which includes hundreds of scientists at UCSF and elsewhere. Because scientists with federal grants cannot pivot quickly to another research area without some other form of financial support, longtime UCSF donor Ron Conway jumped into the breach, directing some of his own funds to QCRG and quickly rounding up a group of other philanthropists, as well as promoting the promising work on social media. To date, more than 40 others have responded with donations to the cause.
With Conway’s support, QCRG scientists, including a group of experts in drug discovery and development, took the new protein map and ran with it, in a search for potential treatments.
One such scientist, Kevan Shokat, PhD, a chemist and expert in drug design, is combing through databases of drug structures to find candidates that match up well with the human host proteins in the SARS-CoV-2 interactome, and therefore might block the actions of the virus. Shokat, UCSF professor of Cellular and Molecular Pharmacology, recruited other labs in his orbit to work on this question.
“We’re kind of crowdsourcing it,” Shokat said. “Everyone is seeing what compounds we have in our labs, work we haven’t published yet, drugs that are already commercially available, or drugs that other people have developed.”
The team is prioritizing promising drugs already approved by the U.S. Food and Drug Administration for any condition, because they could be readily offered to patients with COVID-19. “We’d really like to find a clinically accessible drug.”
Shokat, with fellow QCRG members Brian Shoichet, PhD, professor of Pharmaceutical Chemistry, and Jack Taunton, PhD, professor of Cellular and Molecular Pharmacology, have already shipped promising drug candidates to France’s renowned Institut Pasteur, in Paris, and the Icahn School of Medicine at Mount Sinai in New York, two of the only sites in the world currently working with live cultures of the SARS-CoV-2 virus.
All this work has been done at a breakneck pace, thanks to the immense advances in biological tools and technology over the past decade. When Krogan built a similar host-protein map for HIV in 2012, it took a couple of years.

Nevan Krogan, PhD, talks on a video conference call with members of the QBI Coronavirus Research Group.
“Now we’re replicating that effort for SARS-CoV-2, but in a matter of weeks,” he said. “It’s remarkable, when people are open to collaboration, the speed at which you can get things done.”
All in all, the researchers have identified nearly 70 existing drugs that may be able to disrupt the protein pathways that the new virus exploits.
“We’re digging deep into our current laboratory projects and looking for threads we can add to Nevan’s network,” said Shokat.
One hopeful target is the cellular process known as translation, which viruses hijack to make our cellular machinery crank out their own proteins. Shokat and his QCRG colleague, Davide Ruggero, PhD, the Helen Diller Family Professor in Basic Cancer Research at UCSF, had already been looking for ways to control translation for other diseases. The company eFFECTOR Therapeutics, co-founded by Shokat and Ruggero, has been testing Zotatifin, a drug that inhibits translation, in a Phase 1/2 trial for cancer.
“We can’t wait to see if this drug also works to stop viral replication,” he said.
Gumming up the works of human host proteins hasn’t been the most favored idea in virology, because compromising protein function may produce side effects that take a toll on already vulnerable patients. For dire illnesses like COVID-19, though, Shokat says drugs like Zotatifin can still be a good candidate for therapy because they would only be used short term.
Still, it’s not certain that using existing drugs to immobilize host proteins will be effective and safe for patients sick with COVID-19, making side effects a valid concern.
“These proteins perform normal functions in the host cell besides helping the virus replicate,” said John Young, PhD, global head of infectious diseases at Roche Pharma Research and Early Development in Basel, Switzerland, who has worked with Krogan for several years and is now involved in the QCRG effort. “So, the question is, ‘Can you safely modify the functions of these proteins?’”
Connecting Teams Across the World
Krogan was able to assemble this massive scientific collaboration so quickly because it follows the ambitious interdisciplinary philosophy he has championed at UCSF, and around the world, as the founder of QBI, which is based in the UCSF School of Pharmacy. The deep connections he has built with his “silo-busting” approach are now paying off, he said.
“We’re in a really great situation to bring all these individuals together because we’ve been working with them and making connections amongst them since the inception of QBI three and a half years ago,” Krogan said. “I’ve never seen this many scientists bring their diverse expertise together to work on a single problem,” he says.
The dozens of researchers who joined forces in QCRG have done so, Krogan said, because they believe that despite the issue of potential side effects, targeting host proteins has a lot of benefits over targeting the virus directly.
We’re in a really great situation to bring all these individuals together because we’ve been working with them and making connections amongst them since the inception of QBI three and a half years ago. I’ve never seen this many scientists bring their diverse expertise together to work on a single problem.
One big advantage of the host-directed approach, said Shokat, is that when viruses replicate, they’re highly prone to error. The mutations that arise from those errors are what drives drug and vaccine resistance (thus explaining why you need a new flu shot every year).
“Our own cells’ ways of copying DNA are much more reliable,” he said, “so if we target drugs to our host proteins, we target proteins that are less likely to make changes that are beneficial to the virus.”
It’s also vital to have companies like Roche that are deeply engaged in drug development working with QCRG to provide an industry perspective, Young said.
“Academia and industry are complementary to each other,” he said, “with industry focused on the balance between safety and efficacy of drugs.” That balance, Young pointed out, is of central importance in determining whether or not a drug that works well in a petri dish should be used in a human.
Ultimately, the broad community that Krogan has brought together is carrying out research that will have implications far beyond SARS-CoV-2, which shares characteristics with many related viruses.
“What’s great about targeting these host factors is that if we find a weak link and can target it with a drug, that drug will work on COVID-21 or whatever comes next, and any other virus that uses the same pathway,” he said.
Krogan believes there are indeed weak links, in SARS-CoV-2 infection and in many other diseases, and his interactome maps illuminate them.
“We see proteins and complexes and pathways come up again and again across a variety of diseases,” said Krogan. “Why? I think it’s because there’s a kind of Achilles heel in your cells.” These same pathways also appear to play a role in seemingly unrelated diseases, such as heart disease and cancer.
Krogan said his consortium approach allows researchers to do the essential science of this moment while paving the way for future advances.
“We’re setting up infrastructure to tackle the next pandemic,” he said. “We’re unique in that, here at UCSF, we can do things like this as a group and also pull in talented people from around the world. We’re setting a new paradigm of how science should be done.”